Description
This script reproduces some results of the study of simulated LFPs.
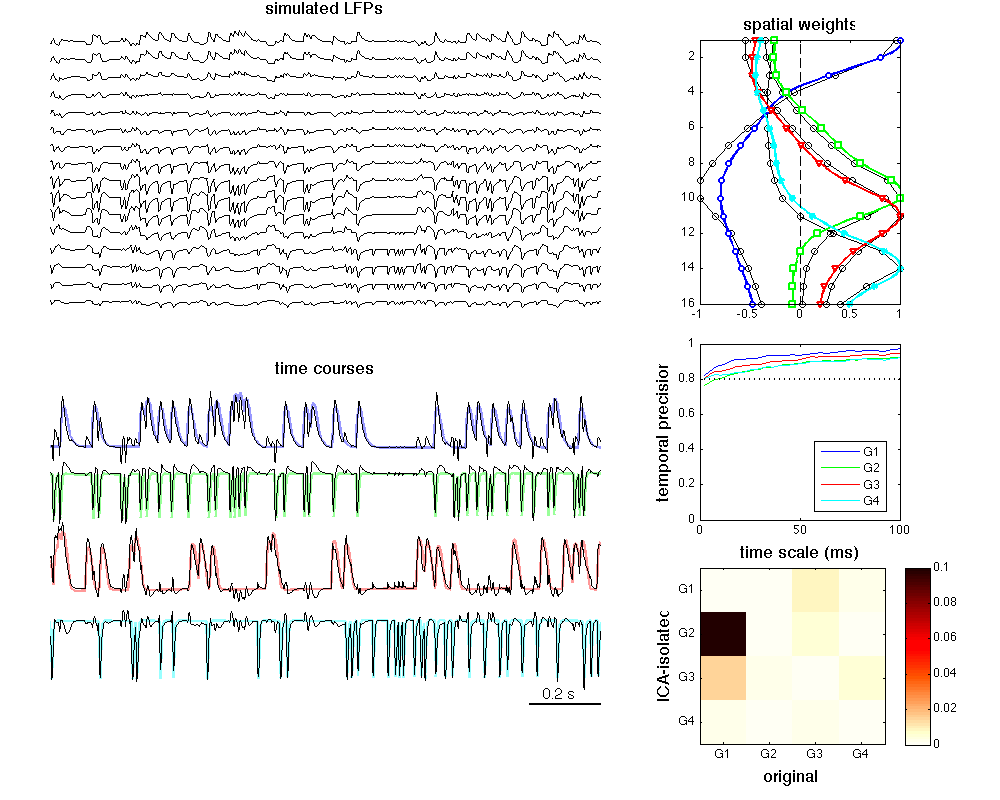
It corrsponds to Fig. 2 for Makarova et al. (2011). Parallel Readout of Pathway-Specific Inputs to Laminated Brain Structures.
Frontiers in Systems Neuroscience.
After reviewer's revision. July 21, 2011.
Contents
Load simulated data
close all clear all cd('/Users/valeri/Dropbox/Julia/Frontiers Neuroscience 2011/Revision'); path(path,'./MatFunc'); path(path,'/Users/valeri/Dropbox/work/LFPanalysis/MatFunc/'); % Original separate generators load('./DATA/G1_ICA.mat'); os = G.s(1,:); oM = G.V(:,1); load('./DATA/G2_ICA.mat'); os = [os; G.s(1,:)]; oM = [oM G.V(:,1)]; load('./DATA/G3_ICA.mat'); os = [os; G.s(1,:)]; oM = [oM G.V(:,1)]; load('./DATA/G4.mat'); os = [os; G.s(1,:)]; oM = [oM G.V(:,1)]; % ICA-isolated LFP-generators load('./DATA/G1+G2+G3+G4.mat'); s = G.s; Fs = Args.Fs; N = size(s,1); t = ((1:size(s,2))-1)/Fs;
Evaluate the Cross-Contamination Index
xShift = 2000; % remove transient part
os = CenterZeroValue(os, xShift);
s = CenterZeroValue(s, xShift);
s = AdjustLoadings(s, os, xShift);
s = CenterZeroValue(s, xShift);
CCind = GeneratorCrossContamination(s(:,xShift:end), os(:,xShift:end));
CCind = CCind - eye(size(CCind));
Evaluate temporal precision
maxScale = 100; wname = 'haar'; ntw = 3; Ratio = 1; scales = 2:2:(maxScale/Ratio); for k = 1:N, WC{k} = wcoher(os(k,xShift:end),s(1,xShift:end),scales,wname,'ntw',ntw); end
Plot Figure 2
XLm = [1 2.55]; h = figure('color','w','position',[100 100 1000 800]); subplot('position',[0.05 0.6 0.55 0.37]) PlotEEG(LFP, Fs, 'e') xlim(XLm) axis off title('simulated LFPs','FontSize',14) subplot('position',[0.05 0.12 0.55 0.4]) Dy = 500; YY = [0 -0.4 -2.2 -2.7]; clr = [0.6 0.6 1; 0.6 1 0.6; 1 0.6 0.6; 0.6 1 1]; hold on for k = 1:N, plot(t, os(k,:) + Dy*YY(k), 'color', clr(k,:), 'LineWidth', 2); plot(t, s(k,:) + Dy*YY(k), 'k') end Dx = 0.1*round(diff(XLm)); ylm = get(gca, 'Ylim'); plot([XLm(2),XLm(2)-Dx],ylm(1)*[1 1],'k','LineWidth',2) text(XLm(2)-Dx*0.8, ylm(1) + 0.03*diff(ylm),[num2str(Dx) ' s'],'FontSize',12) xlim(XLm) title('time courses','FontSize',14) axis off subplot('position',[0.7 0.62 0.2 0.33]) plot(G.V, 1:16,'ko-') hold on PlotLoadingsYX(oM) xlim([-1 1]) title('spatial weights','FontSize',14) subplot('position',[0.7 0.35 0.2 0.22]) clr = 'bgrcmyk'; for k = 1:N plot(Ratio*scales,mean(abs(WC{k}),2),[clr(k) '-'], 'LineWidth',1); hold on end plot(Ratio*[scales(1), scales(end)],0.8*[1 1],'k:','LineWidth',2); legend({'G1','G2','G3','G4'},'location','best') axis([0 scales(end)*Ratio 0 1]) xlabel('time scale (ms)','FontSize',14) ylabel('temporal precision','FontSize',14) subplot('position',[0.7 0.07 0.25 0.22]) imagesc(CCind, [0 0.1]) colorbar m = zeros(64,3); m(:,1) = 1.1 - ((1:64) - 35)/30; m(:,2) = 1 - ((1:64) - 5)/30; m(:,3) = 1 - (1:64)/25; m(m > 1) = 1; m(m < 0) = 0; colormap(m) ylabel('ICA-isolated','FontSize',14) xlabel('original','FontSize',14) set(gca,'YTick',1:N); set(gca,'YTickLabel',{'G1','G2','G3','G4'}) set(gca,'XTick',1:N); set(gca,'XTickLabel',{'G1','G2','G3','G4'})