
 |
 |
 |
 |
 |
 |
 |
 |
|
Ana Carpio, Applied Mathematics - Biological Sciences
We are
developing multiscale models to describe cellular
dynamics and interaction with the environment. Applied
to biofilm formation, we have been able to explain the
emergence of filamentary structures in medical flow
circuits using discrete rod models, as well as
wrinkled patterns on surfaces exploiting Foppl-Von
Karman descriptions for sheets. Coupling with models
of cellular metabolism fitted to data on the action of
drugs, we have been able to simulate the action of
antibiotics on their structure. More recently, we have
applied topological data analysis to the study of gene
expression in cancer processes and to the automatic
characterization of cellular/material interfaces. We
are using clustering techniques to explore data from
immune disorders.
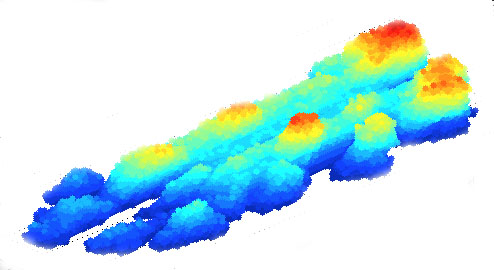
Biofilm
patterns
Previous work in this field
fitted oscillator models to describe quantitatively
and qualitatively recent experiments of protein
unfolding and refolding by means of wavefront
analysis. Earlier, we characterized propagation
failure of nerve impulses along myelinated nerves in
discrete Fitz-Hugh Nagumo and Hodgkin-Huxley type
models by a similar discrete wavefront analysis.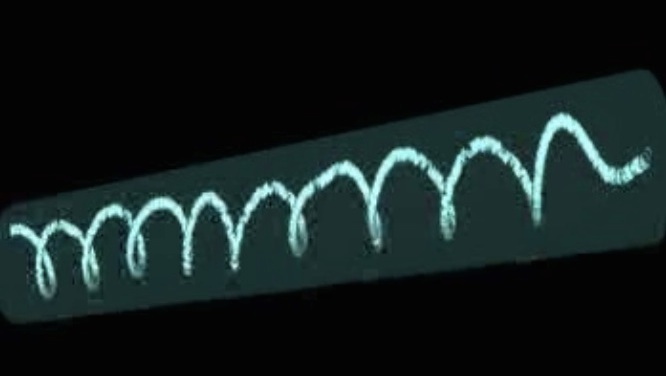
|
 |
 |
| Helical biofilm wrapping around tube walls in a laminar flow past a stenosis (movie) | Succesive
wrinkle branching in
a biofilm growing on agar (movie) |
Biofilm streamer
formation and detachment in straight ducts (movie) |
- Pattern
recognition in
data as a diagnosis
tool
(with A.
Simón, A.
Torres, L.F.
Villa),
Journal of
Mathematics in
Industry 12,
3, 2022 [pdf]
[arxiv]
- Positivity preserving high order schemes for kinetic models of angiogenesis (with E. Cebrián), International Journal on Nonlinear Sciences and Numerical Simulation, 2022 to appear [pdf] [arxiv]
- Immersed boundary approach to biofilm spread on surfaces (with R. González-Albaladejo), Communications in Computational Physics 31(1), 257-292, 2022 [pdf] [arxiv]
- Parameter identification in epidemiological models (with E. Pierret), in Mathematical Analysis of Infectious Diseases (MAID 2020), P. Agarwal, J.J. Nieto, D.F.M. Torres, Eds., Chapter 7, 103-124, Elsevier 2022 [pdf]
- Uncertainty
quantification
in covid19
spread:
lockdown
effects (with
E. Pierret),
Results
in Physics 35,
105375, 2022 [pdf]
[arxiv]
- Mathematical
models of the
spread and
consequences
of the SARS-CoV-2
pandemics:
Effects
on health,
society,
industry,
economics and
technology
(with A. Micheletti,
A. Araújo, N.
Bubko, M.
Ehrhardt), Journal
of Mathematics
in Industry
11, 15, 2021 [pdf]
- Clustering methods and Bayesian inference for the analysis of the time evolution of immune disorders (with A. Simón, L.F. Villa), preprint 2020 [arxiv]
- Tracking
collective
cell motion
by topological
data analysis
(with L.L.
Bonilla, C.Trenado),
PLOS
Computational
Biology
16(12),
e1008497, 2020
[pdf][arxiv]
- Incorporating cellular stochasticity in solid-fluid mixture biofilm models (with E. Cebrián), Entropy 22(2), 188, 2020 [pdf]
- Fingerprints of cancer by persistent homology (with L.L. Bonilla, J.C. Mathews, A.R. Tannenbaum), bioRxiv 777169, 2019 [pdf]
- Biofilms as poroelastic materials (with E. Cebrián, P. Vidal), International Journal of Non-linear Mechanics 109, 1-8, 2019 [pdf]
- Dynamics of Pseudomonas putida biofilms in an upscale experimental framework (with D.R. Espeso, E. Martínez-García, V. de Lorenzo), Journal of Industrial Microbiology & Biotechnology 45, 899-911, 2018 [pdf]
- Biofilm mechanics and
patterns
(with E. Cebrián, D. R.
Espeso, P. Vidal), in
Coupled Mathematical
Models for Physical
and Biological
Nanoscale Systems and
Their Applications, Springer
Proceedings in Mathematics
& Statistics 232,
Springer Nature 2018, [pdf]
- Dynamic energy budget approach to evaluate antibiotic effects on biofilms (with B. Birnir, E. Cebrián, P. Vidal), Communications in Nonlinear Science and Numerical Simulation 54, 70-83, 2018 [pdf] [arxiv]
- Stenosis triggers spread of helical Pseudomonas biofilms in cylindrical flow systems (with D.R. Espeso, E. Martínez-García, V. de Lorenzo), Scientific Reports 6, 27170, 2016 [pdf]
- Dynamics of bacterial
aggregates in microflows (with
B. Einarsson, D.R. Espeso), in
Progress in
Industrial Mathematics at
ECMI 2014, Mathematics in Industry 22, Springer 2016 [pdf]
- Differential growth of wrinkled biofilms (with D.R. Espeso and B. Einarsson), Physical Review E 91(2), 022710, 2015 [pdf] [arxiv]
- Theory of force-extension curves for modular proteins and DNA hairpins (with L.L. Bonilla and A. Prados), Physical Review E 91(5), 052712, 2015 [pdf] [arxiv]
- Protein unfolding and refolding as transitions through virtual states (with L.L. Bonilla and A. Prados), EPL (Europhysics Letters) 108(2), 28002, 2014 [pdf] [arxiv]
- Sawtooth patterns in force-extension curves of biomolecules: an equilibrium-statistical-mechanics theory (with A. Prados and L.L. Bonilla), Physical Review E 88(1), 012704, 2013 [pdf] [arxiv]
- Biofilm growth on rugose surfaces (with D. Rodriguez, B. Einarsson), Physical Review E 86(6), 061914, 2012 [pdf] [archivo]
- Spin-oscillator model for the unzipping of biomolecules by mechanical force (with A. Prados, L.L. Bonilla), Physical Review E 86(2), 021919, 2012 [pdf] [arxiv]
- Propagation failure along myelinated nerves (with I. Peral), Journal of Nonlinear Science 21(4), 499-520, 2011 [pdf] [archivo]
